Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), responsible for coronavirus disease 19 (COVID-19), has emerged in December 2019 when the first case was reported in Wuhan, China. Soon after, it has rapidly spread to other countries worldwide, becoming pandemic with more than 4 million fatalities and 230 million cases registered1.
There are different factors that make it difficult to contain the spread of COVID-19. These include the high mutation rate of the virus, the challenge of diagnosing asymptomatic or mildly symptomatic individuals and the capability of the virus to be transmitted during the pre-symptomatic phase2.
After transmission processes through respiratory droplets, aerosol or surface contamination, follows the incubation period that could led to a plethora of symptoms such as fever, cough, shortness of breath, loss of taste and smell, diarrhea and nausea3. Nevertheless, a notable proportion of individuals with pre-existing conditions, such as asthma, diabetes, cardiovascular disease and other chronic illnesses experienced severe complications such as pneumonia affections or acute respiratory syndrome4. Some respiratory failures in severe SARS-CoV-2 infection have been found to be associated with the activation of immune response and pro-inflammatory mechanisms by chemokines and cytokine release, which may be caused by a “cytokine storm syndrome”5. In addition to pre-existing clinical conditions, other factors, such as age, sex, and ethnicity can also impact the clinical presentation of infected patients6.
As the vast range of disease susceptibility and outcomes observed in individuals infected with SARS-CoV-2 may be attributed to gene expression modulation resulting from virus-host cell interactions, several studies have been performed to investigate the biological effects of virus infection on the host transcriptome profile7. SARS-CoV-2 enters into the host cell by direct attachment to multiple receptors on the cell membrane or through membrane fusion within the endosome after endocytosis leading to further factors in human gene expression modulation8. SARS-CoV-2 primarily enters host cells through the angiotensin converting enzyme-2 (ACE2) located on the surface of different cell types. This interaction activates the renin-angiotensin pathway, which may increase the risk of severe COVID-19 symptoms in affected individuals9. Hence, upon detection of infection, human cells activate mechanisms to counteract viral replication which involves significant reprogramming of their own transcriptome10. Despite the worldwide spread, the host immune response against SARS-CoV-2 infection remains poorly characterized. Identifying transcriptome differences can be valuable for the determination of the cellular pathways that are modulated by the virus in infected cells.
Here, our objective is to provide a comprehensive transcriptomic dataset of a cohort of SARS-CoV-2 positive Italian individuals. This dataset will allow the scientific community to study the impact of virus infection on the transcriptome of mucosa cells. To this aim, RNA extracted from 35 nasopharyngeal swabs of COVID-19 patients enrolled in the Campania region was subjected to total RNA sequencing and subsequent bioinformatics analysis (Fig. 1).
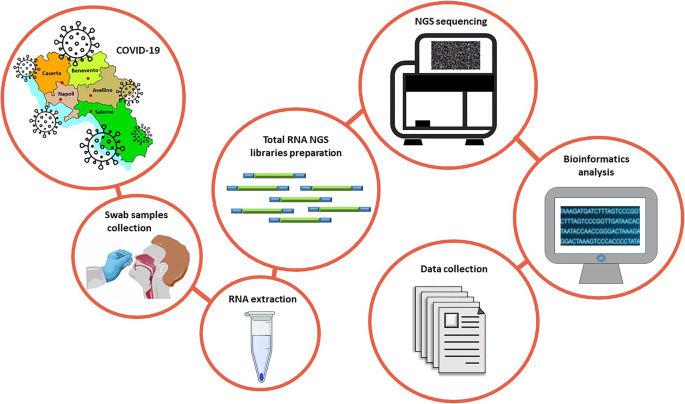
Experimental workflow. Summary of the experimental workflow applied to generate transcriptomic datasets originated by RNA extracted from nasopharyngeal swabs in the Campania cohort.
Patients were selected according to age, sex, sampling time and clinical manifestation of the disease (Fig. 2a and Supplementary File 1). Our sampling also covers the timing of the three different waves of SARS-CoV-2 infections in Italy, ranging from the pandemic declaration in March 2020 to spring 202111. In detail, 15 cases belong to the 1st period (March-May 2020), 13 to the 2nd period (September – November 2020) and 7 to the 3rd period (January – February 2021) (Fig. 2a).
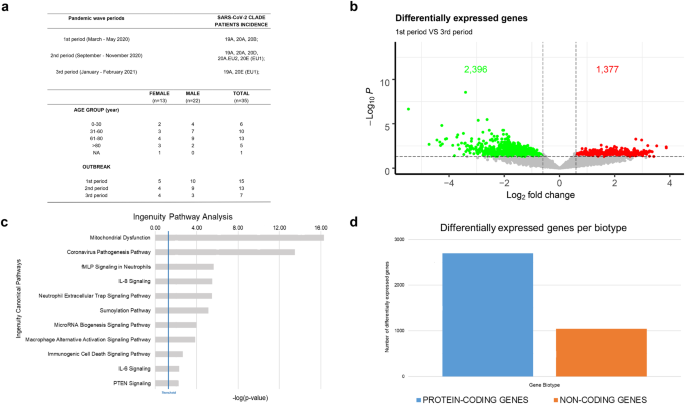
Analysis of RNA gene expression profiles between 1st and 3rd wave (a) Summary of demographics and other patients’ features of recruited cases in this cohort. (b) Volcano plot summarizing transcripts changes comparing 1st vs 3rd period. Green Dot and Red Dot show down- and up-regulated genes, respectively. According to the adjusted p-values (FDR) threshold of 0.05, transcripts associated with insignificant expression values are reported in grey. The dotted line (threshold) represents the cut-off (p-value ≤ 0.01). (c) Bar chart showing statistically significant pathway, according to IPA, of differentially expressed genes between 1st vs 3rd pandemic waves in Campania. (d) Bar chart showing the differentially expressed genes biotype detected in the analysis, divided into protein-coding (blue) and non-coding (orange) genes.
Interestingly, by total RNA approach and deep sequencing conditions, detailed in the Methods section, our bioinformatics analyses have detected also reads aligned on the SARS-CoV-2 genome. In this way, we are also able to observe the distribution of virus variants peculiar to different pandemic waves, which may contribute to host response variability analysis (Fig. 2a and Supplementary File 1).
The transcriptome dataset here proposed, can provide valuable insights into the biological impact of SARS-CoV-2 infection on the modulation of host gene expression. By analyzing this dataset and integrating it with others, researchers could identify key protein-coding and non-coding genes involved in pathways affected by the virus’s entrance. This could help in the development of new therapies and diagnostic tools.
Moreover, this dataset includes several clinical factors which can be used to study the relationship between these factors and the host’s gene expression changes induced by SARS-Cov-2 infection.
Additionally, the clade assignment provides an opportunity to investigate the potential differences in transcriptome profiles between different viral strains. This can help in understanding the pathogenesis of the disease and the potential differences in virulence and transmissibility among different SARS-CoV-2 variants. Overall, the transcriptome dataset from the Italian cohort of these patients is a valuable resource for researchers to be integrated with other datasets and identify potential therapeutic targets and diagnostic biomarkers.
